Acknowledgment
I am grateful to Moritz Wette for working through this tutorial and checking its consistency
Table of Contents
To download mrtailor, please return to the mrtailor main page.
- Data used in this tutorial
- Step 1: Preparing the multiple alignment file with blast
- Step 2: A simplified molecular replacement with 1DBI
- Step 3: Direct refinement with refmac5
- Step 4: Refinement with external restraints from prosmart
- Step 5: Refinement with external restraints after using mrtailor
- Step 6: Refinement with improved input model
- Step 7: Comparing results
mrtailor Tutorial
This tutorial for the program mrtailor is based on structural data of "tripeptidyl-peptidase I" (TPP) ([Pal et al., 2009], PDB-ID 3ee6).
Notation: Any line in this tutorial starting with
#>
marks a command which should be typed at a terminal.
The Data
In order to follow the tutorial, you should install mrtailor and download the following files (there will be appropriate link when each file is required, and you do not need to download all files now):- TPP sequence tpp.fasta in fasta format.
- Data file tpp_tutorial_data.mtz
- PDB file tpp_tutorial_data.pdb. It corresponds to the PDB ID 3EE6 [Pal et al., 2009] of the target structure.
- PDB file 1dbi.pdb [Smith et al., 1999].
Step 1: Blast
In this section we carry out a Blast search in order to find a PDB file which putative similarity to the structure of TPP, and create the alignment file which will be used by Mr Tailor later on.
The first step to find a candidate for external restraints is a Blast Search against the PDB:
- Paste the sequence file tpp.fasta into the sequence field.
- Select the "Protein Data Bank proteins (pdb)" as database!
- Hit the "BLAST" button at the bottom of the page!
- Of course the list of hits includes the deposited 3EE6.pdb. Since it is more realistic, choose the PDB file 1DBI which covers only 52% of the sequence with a maximum identity of 13%.
- At the "Sequences producing significant alignments with E-value BETTER than threshold" listing of sequences, select "All" and click on the "Multiple Alignment" link just below the section title!
- This openes a new tab to the Cobalt tool. Click on "Download" and select "Clustal Download alignment". This offers you a file with a slightly cryptic name. As this tutorial is written, it is called "NBBVUW4G211-alignment.aln". Once downloaded, rename it to "tpp_blast.aln".
The blast alignment file contains the GenInfo Sequence Accession Number rather than the original PDB codes. You would have to count the lines on the Cobalt web site that the sequence for 1DBI has the name gi|6573500 (number 13) in the file "tpp_blast.aln"
The first sequence gi|215261288 corresponds to 3EE6.
Step 2: Preparing for Refinement
In this section we mimic a molecular replacement solution - Mr Tailor is primarily meant for better external restraints, so an actual structure solution for the TPP data is not the purpose of this tutorial.
- Start the program Coot [Emsley et al., 2010].
- Load the target PDB file for TPP, tpp_tutorial_data.pdb
- and load the template PDB file 1dbi.pdb! (you could also use File -> Fetch PDB using Accession Code from the coot menu).
- The "layz molecular replacement" consists of
Calculate -> SSM Superpose in Coot: Make sure to choose chain A of 3ee6 as "Reference
Structure" and chain A of 1dbi as "Moving Structure".
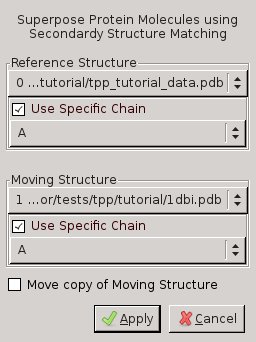
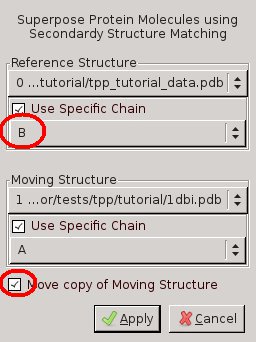
Figure 1: Screenshot of Coot's SSM Superposition Menu. Left: chain A of 1DBI onto chain A of 3EE6. Right: chain A of 1DBI onto chain B of 3EE6 - TPP is a homodimer, but the structure 1DBI contains a monomer. Therefore repeat the previous step, but this time select chain B for tpp_tutorial_data.pdb and click "Move copy of Moving Structure" in the SSM Superpose Menu!
- From the Coot main menu select: Calculate->Merge Molecules ... the second instance of 1dbi (Copy_of_1dbi.pdb Chain A) into 1dbi.pdb.
- Save the coordinate of the now dimer 1dbi.pdb as 1dbi_ssm-AB.pdb
I recommend removing all molecules and reloading the newly created PDB file 1dbi_ssm-AB.pdb to ensure it really represents the dimer in place of the two chains for TPP-I.
Tidying up the PDB file 1dbi_ssm-AB.pdb
Remove all water molecules from the new file and insert the correct space group and cell (otherwise, refmac5 [Murshudov et al., 2011] complains) with the CCP4 program pdbset. The instructions
exclude hetero
exclude water
cell 113.450 128.930 100.500 90.00 90.00 90.
space P21212
end
are found in the script pdbset.script and
called via
#> pdbset XYZIN 1dbi_ssm-AB.pdb < pdbset.script
#> mv XYZOUT 1dbi_ssm-AB.pdbThe second command renames the output file XYZOUT to 1dbi_ssm-AB.pdb.
The resulting file also contains three Ca atoms and a Na atom, and you need a text editor to remove those lines near the end of chain A and near bottom of the file (This is not essential, but it makes sense to mimic a realistic case).
Step 3: Running Refmac5 [Murshudov et al., 2011]
Refmac is run from the command line using the script refmac_pure.sh. This is a script for low resolution refinement with a low matrix weight (weight MATRIX 0.005) and a large number of cycles (ncyc 100)
#> bash refmac_pure.sh | tee refmac_pure.logrefmac5 is going to run for about half an hour or more, so continue with the next step.
Step 4: Running Refmac5 with ProSmart [Nicholls et al., 2012]
While refmac5 is running, open a new terminal to continue with the tutorial!
At this stage, the input file for refmac5 has not really changed because there has not yet been any refinement, but we are still going to use 1DBI as reference file for prosmart:#> prosmart -p1 1dbi_ssm-AB.pdb -p2 1dbi.pdb -o prosmart
This creates the file ./prosmart/1dbi_ssm-AB.txt.txt containing external restraints for refmac5, which is run with the script refmac_prosmart.sh:
#> bash refmac_prosmart.sh | tee refmac_prosmart.log
As you compare the two refmac-scripts you will notice the extra lines
external weight scale 500
@./prosmart/1dbi_ssm-AB.txt
Check the log file refmac_prosmart.log a few minutes after starting refmac5 to notice the listing of external restraints:
Standard External All
Bonds: 7862 23442 31304
Angles: 14170 0 14170
Chirals: 644 0 644
Planes: 1324 0 1324
Torsions: 3220 0 3220
while the same table for the refmac_pure-run contains zero external restraints.
Step 5: MrTailor
First create the clustalx scores file from the alignment file:
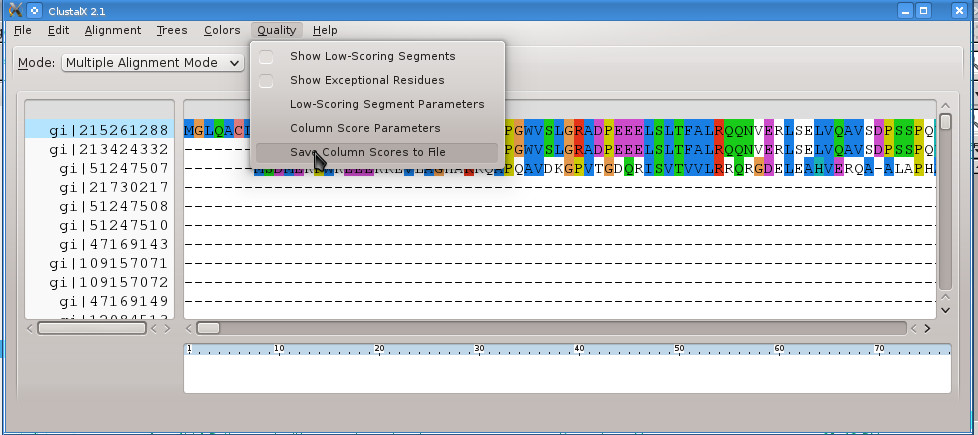
#> clustalx tpp_blast.aln
Click on the top sequence name gi|215261288 which is the sequence of TPP-I (Unfortunately the Blast download file replaces PDB IDs with their "gi" ID)! Select Quality -> Save Column Scores to File and save it to tpp_blast.qscores!
Start the mrtailor-gui with the command
#> mrtailor-gui &
and fill in the fields as shown in Figure 2!
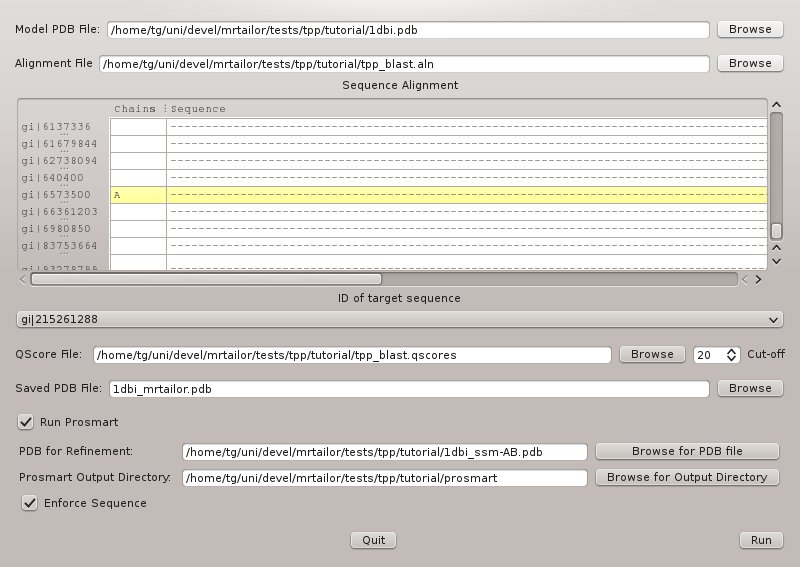
- Model PDB: 1dbi.pdb
- Alignment File: tpp_blast.aln
- ID of target sequence: gi|215261288
- QScore File: tpp_blast.qscores
- Output PDB File: 1dbi_mrtailor.pdb
- PDB for Refinment: 1dbi_ssm-AB.pdb
- Prosmart Output Directory: prosmart_output
The corresponding command line reads
#> mrtailor -a tpp_blast.aln -m "gi|6573500" -p 1dbi.pdb -t "gi|215261288" \
-o 1dbi_mrtailor.pdb -q tpp_blast.qscores -r 1dbi_ssm-AB.pdb -o prosmart
Clicking 'Run' actually results in the error message Figure 4!

Since the file 1dbi_ssm-AB.pdb is the molecular replacement solution, it does not contain the sequence for 3EE6, but 1DBI and hence does not match the target sequence. How to proceed?
Mapping the Sequence of TPP-I
mrtailor can be used to map the sequence of 3EE6 onto the structure of 1DBI. In order to do so, use the input as shown in Figure 5.
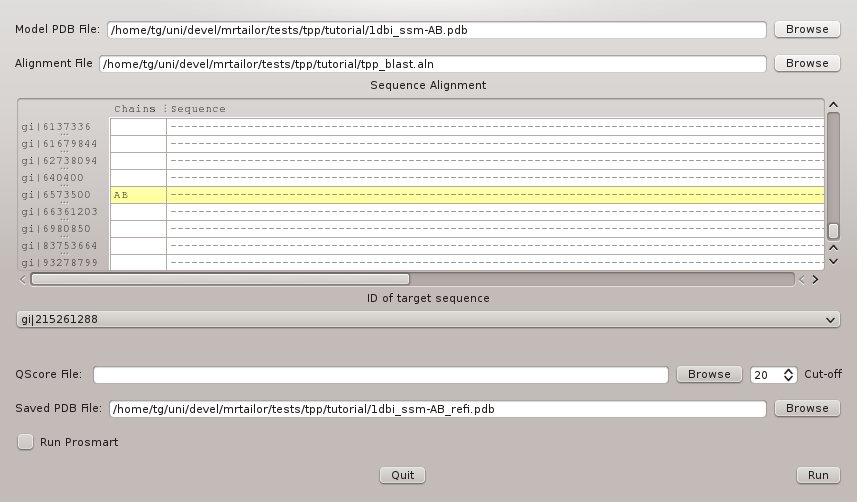
The corresponding command line reads:
#> mrtailor -a tpp_blast.aln -t "gi|215261288" -m "gi|6573500" -p 1dbi_ssm-AB.pdb -o 1dbi_ssm-AB_refi.pdb
Corrected Run of mrtailor
Next run mrtailor again with the generated PDB-file 1dbi_ssm-AB_refi.pdb as refinement input. The configuration is displayed in Figure 6, and the command line reads:
#> mrtailor -a tpp_blast.aln -t "gi|215261288" -m "gi|6573500" -p 1dbi_ssm-AB.pdb -o
1dbi_mrtailor_corrected.pdb -q tpp_blast.qscores -r 1dbi_ssm-AB_refi.pdb -o prosmart
mrtailor will run prosmart separately for each chain found in 1dbi_ssm-AB_refi.pdb; in this particular case with the PDB file being a homo dimer, this is equivalent to calling prosmart as
#> prosmart -p1 1dbi_ssm-AB_refi.pdb -p2 1dbi_ssm-AB.pdb
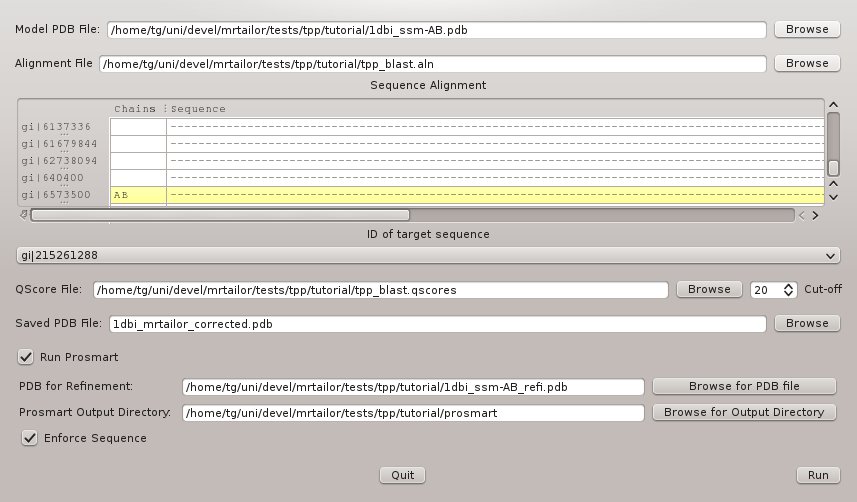
The corresponding command line reads
#> mrtailor -a tpp_blast.aln -m "gi|6573500" -p 1dbi_ssm-AB.pdb -t "gi|215261288" \
-o 1dbi_mrtailor_corrected.pdb -q tpp_blast.qscores -r 1dbi_ssm-AB_refi.pdb -o prosmart
If the template were a PDB file consistent of several different subunits, the result would, however, be different. Therefore, the GUI has created separate output directories for each chain matching the target sequence gi|215261288:
The corresponding lines for the script to run refmac5 are:
external weight scale 500
@./prosmart_chain_A/1dbi_ssm-AB_refi.txt
@./prosmart_chain_B/1dbi_ssm-AB_refi.txt
Step 6: Running Refmac5 with Mr Tailor's PDB file
Download the script refmac_mrtailor.sh and run it:
#> bash refmac_mrtailor.sh | tee refmac_mrtailor.log
The number of external restraints is now lower than from 1dbi.pdb because of the gaps introduced by mrtailor:
Standard External All
Bonds: 3784 14292 18076
Angles: 6250 0 6250
Chirals: 402 0 402
Planes: 822 0 822
Torsions: 1376 0 1376
Step 7: Results
At low resolution R and Rfree values can be very high, and they do not necessarily make a good criterium whether or not the solution is correct. E.g. even though from the fake molecular replacement applied in this tutorial, the R and Rfree values after the first round of refinement are above 50%:
| pure | prosmart | mrtailor | ||
|---|---|---|---|---|
| R | init | 55.7% | 55.7% | 56.4% |
| final | 51.8% | 50.4% | 52.9% | |
| Rfree | init | 55.3% | 55.3% | 56.7% |
| final | 55.3% | 54.0% | 55.1% |
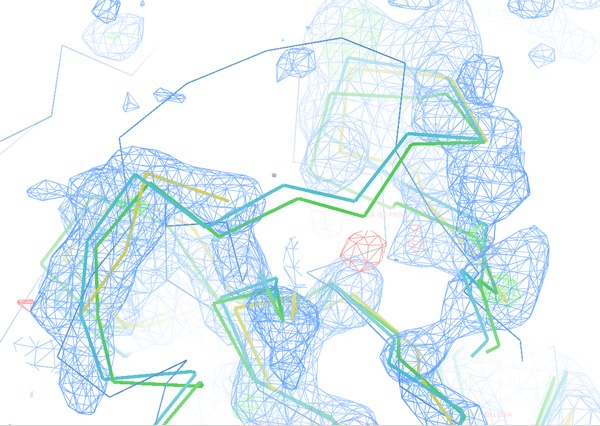
Figure 7 shows a helix between residues A334 and A353 (w.r.t. the original 3EE6 PDB file for TPP-I). The fragmentation of the input PDB-file from refinement after mrtailor allows this fragment (green fragment) to shift towards the correct coordinates (grey fragment) compared to the original PDB-file with or without external restraints (red and orange fragments). The rmsd for 16 Cα atoms with respect to the (grey) target coordinates are:
mrtailor (green): 1.09 Å, prosmart (red) : 2.02 Å, and pure (no external restraints, orange): 1.80 Å

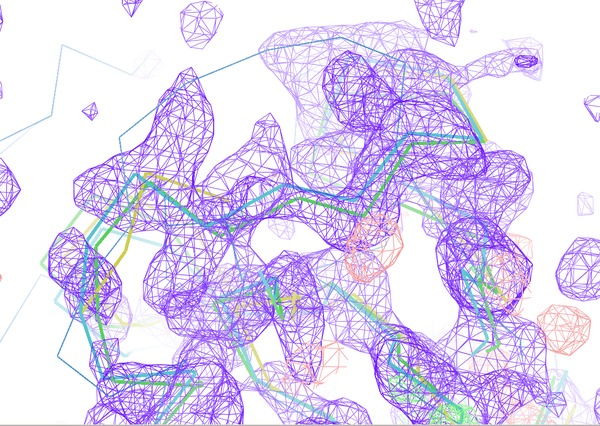
The next Figure 8 compares the electron density map near residue
A266 (w.r.t. the original 3EE6 PDB file for TPP-I).
The map on the left (after using mrtailor) show much weaker model bias towards the course of
the loop of 1DBI. The actual loop of 3ee6 is shown as thin Cα-trace. In such a case it is
more likely that model bias can be removed during model building.
References
- Pal, A. et al. "Structure of Tripeptidyl-peptidase I Provides Insight into the Molecular Basis of Late Infantile Neuronal Ceroid Lipofuscinosis" J. Biol. Chem. (2009), 284: 3976-3984
- M. D. Winn et al. "Overview of the CCP4 suite and current developments" Acta. Cryst. D67, 235-242 (2011)
- Murshudov, G. N. et al., "REFMAC5 for the Refinement of Macromolecular Crystal Structures", Acta Crystallogr. D67 (2011), 355-367
- Nicholls, R. A. et al. "Low-resolution refinement tools in REFMAC5", Acta Cryst D68 (2012), 404-417
- C. A. Smith et al. "Calcium-mediated thermostability in the subtilisin superfamily: the crystal structure of Bacillus Ak.1 protease at 1.8 A resolution.", J. Mol. Biol. (1999), 294, 1027-1040.
- Emsley, P. et al., "Features and Development of Coot", Acta Crystallogr. D66 (2010), 486-501.
Tim Gruene
Last modified: Mar 25, 2020 22:40


