Note: this abstract has been submitted as an abstract for the OHBM2019 meeting in Rome, Italy.
I am interested in collaborations on this project. If you are interested and have experience with JavaScript/Node.js/Vue.js, please get in touch with me.
This is a current development snapshot of our project. Contact me if you intend to use it in your lab to get the latest updates.
Introduction. A ubiquitous challenge for neuroimaging and other forms of empirical research is the organization and quality assessment of collected data. Nowadays there are excellent standards for organizing datasets, such as BIDS (Gorgolewski et al., 2017)and the OpenfMRI format (Poldrack et al., 2013). These structured datasets are optimized for machine-processability, enabling the use of automated scripts for safe and secure data storage, efficient data processing using computational pipelines, and intuitive collaboration between labs. However, before data actually conforms with the a prioridefined syntax and semantics, data needs to be manually transferred and rearranged in an unstructured and insecure fashion across different systems and air-bridged devices, such as an MRI scanner, lab bench, and online questionnaire servers (Figure 1A). Typical solutions for this problem are (a) relying that all lab members are able to responsibly organize their data independently (the hoping for the bestapproach), (b) all data is handed over to one or a few data science experts who are responsible for data management and assessment (the hoping that they will never leave the labapproach), and (c) hybrid approaches where data experts define strict policies on how to organize the data and expect all members (including those with different project requirements) to follow these guidelines (the creating lots of frustration on both sidesapproach). To a different degree, all of these options entail problems such as potential data loss, hard to manage data security and backup strategies, and storage policies that are too inflexible to be applicable for all types of studies. In interdisciplinary teams, it cannot be expected that the required data management skills and coding competences are present in all lab members. This applies in particular for, e.g., the new lab member with a background in molecular psychiatry or the neurophenomenology full professor who cannot be expected to run obscure bash scripts on the lab server to monitor the project’s progress. This was the motivation for developing sweetDatathat provides a user-friendly, efficient, modular, and open framework for management of raw source data.
Methods. sweetData is being developed in JavaScript using Node.js (v8.11.1, Node.js Foundation) featuring a server module based on Express.js (v4.16.4) and a customizable front-end implemented in Vue.js (v2.5.17) with HTML/CSS. sweetData’s server component is a stand-alone webserver responsible for validating the project’s files and folder structure and reporting its status (Figure 1B). The sweetData client (or any other client application, Figure 2A) can access the server-sided service via an API, allowing for a scalable, flexible, and future-proof architecture. The project semantics, i.e., the way files should be organized within a project, are defined using a customizable JSON file (Figure 2B/C).
Results. Currently, sweetData supports the management of text, DICOM, and NIFTI files. A development snapshot of sweetData is provided online (http://homepage.univie.ac.at/ronald.sladky/wp/sweetdata/) and collaboration in this project, in particular to add new data formats (e.g., EEG data, MAT files) is highly encouraged.
Conclusions. With an ever-increasing number of files and heterogeneous data sources, robust and practicable solutions for project data management are highly relevant. Standardized fMRI reporting formats and international collaborations require the use of structuralized and reproducible forms of data management. While software exists for validating if a project conforms to BIDS, a more general form of data validation to customized schemas optimized for source data has been missing. sweetData can provide an interface to enable translating heterogenous forms of source data into a self-defined, well-ordered, structured project format. Finally, these datasets can be converted to other standardized data management schemas.
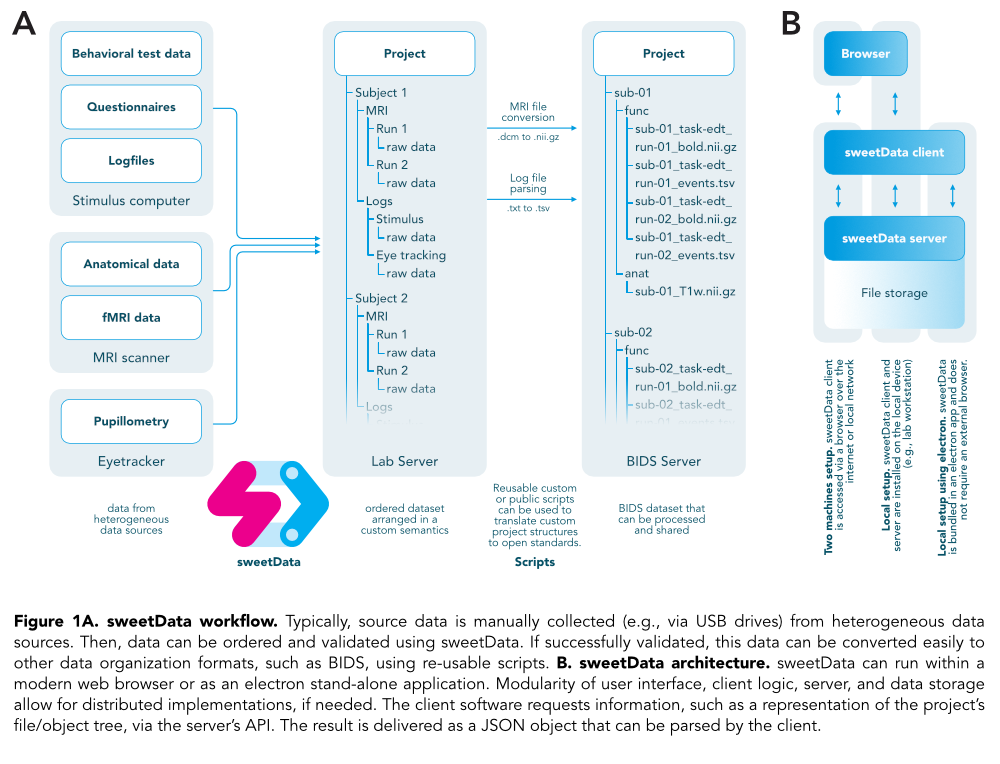
Figure 1A. sweetData workflow.Typically, source data is manually collected (e.g., via USB drives) from heterogeneous data sources. Then, data can be ordered and validated using sweetData. If successfully validated, this data can be converted easily to other data organization formats, such as BIDS, using re-usable scripts. B. sweetData architecture.sweetData can run within a modern web browser or as an electron stand-alone application. Modularity of user interface, client logic, server, and data storage allow for distributed implementations, if needed. The client software requests information, such as a representation of the project’s file/object tree, via the server’s API. The result is delivered as a JSON object that can be parsed by the client.

Figure 2A. sweetData user interface. The user interface is implemented in JavaScript, Vue.js framework, HTML, and CSS and can be easily adapted to new use cases, such as displaying new data formats. Identified entities and validation errors (e.g., missing folder, filesize to small), warnings (e.g., missing optional entity) or notes (e.g., unknown/unnecessary file) are highlighted in the project tree. B Example project config file.A JSON file is used to store the relationships of different entities within a project. In this example, it is assumed that a project rootcontains at least one Subject(e.g., a folder named clamy17-e01), which must contain the folders OpenNFT,MRI, DRIN, and Logs. MRI, in turn, contains different types of NIFTI files, which have a minimum file size and a minimum number of volumes.C Graph of the relationships described in the JSON file.
References
Gorgolewski, K.J., Alfaro-Almagro, F., Auer, T., Bellec, P., Capota, M., Chakravarty, M.M., Churchill, N.W., Cohen, A.L., Craddock, R.C., Devenyi, G.A., Eklund, A., Esteban, O., Flandin, G., Ghosh, S.S., Guntupalli, J.S., Jenkinson, M., Keshavan, A., Kiar, G., Liem, F., Raamana, P.R., Raffelt, D., Steele, C.J., Quirion, P.O., Smith, R.E., Strother, S.C., Varoquaux, G., Wang, Y., Yarkoni, T., Poldrack, R.A., 2017. BIDS apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Comput Biol 13, e1005209.
Poldrack, R.A., Barch, D.M., Mitchell, J.P., Wager, T.D., Wagner, A.D., Devlin, J.T., Cumba, C., Koyejo, O., Milham, M.P., 2013. Toward open sharing of task-based fMRI data: the OpenfMRI project. Front Neuroinform 7, 12.
